Professor of Genetic Epidemiology and Statistical Genetics, University of Edinburgh.
also
For the Usher Institute, information about research is on this page, information about taught postgraduate courses is on this page, and information about PhDs is on this page. I am a listed supervisor for the UKRI Centre for Doctoral Training in Biomedical Artificial Intelligence, which offers a four-year training program with the first year spent working for a Master’s degree through taught courses and projects, followed by a three-year PhD programme.
In the Usher Institute I am a member of the Molecular Epidemiology Group. I have a secondary affiliation to the Centre for Genomic and Experimental Medicine in the Institute of Genetics and Molecular Medicine.
From 2019 to 2022 I was on the Medical Research Council’s Methodology Research Programme (now renamed Better Methods Better Research) panel. I continue to review grants for UKRI.
Usher Institute
College of Medicine and Veterinary Medicine
University of Edinburgh
Usher Building, 5 Little France Rd, Edinburgh EH16 4UX
Phone +44 131 650 4556
![]()
My research focuses on methods for molecular and genetic epidemiology, with applications in clinical prediction and personalized medicine. These methods make use of Bayesian and computationally-intensive statistical methods, and machine learning methods for constructing predictors. I work closely with Helen Colhoun’s research group at the Centre for Genomic and Experimental Medicine in the Institute of Genetics and Molecular Medicine. This collaboration includes the development of an analysis platform based on deidentified electronic health records and the use of this platform to study drug safety and complications of diabetes.
Since mid-2022 my research has refocused on developing and applying a novel method of genetic analysis that identifies “core” genes for disease by adding up the predicted trans- effects of all SNPs on the expression of each gene, and testing for association with the disease or trait under study. This is based on a fundamental rethink of the genetic architecture of complex traits, first proposed as the “omnigenic” model (Boyle, Li and Pritchard 2017) which postulates that the polygenic effects of common SNPs on a typical complex trait coalesce via trans- effects on the expression of a relatively sparse set of effector genes. We call this the “sparse effector” hypothesis: a more precise term than “omnigenic model”. I am working on this approach with Athina Spilopoulou (Arthritis UK Career Development Fellow), Andrii Iakovliev (MRC Cross-Disciplinary Resarch Fellow), Svitlana Braichenkno (MRC Cross-Disciplinary Research Fellow, Xuan Zhou (post-doc) and Jay Dyer (post-doc). Our group includes two PhD students that I co-supervise.
We have published five papers demonstrating the application of this approach to four immune-mediated inflammatory diseases: type 1 diabetes (Iakovliev et al 2023, Zhou et al 2025), rheumatoid arthritis, systemic lupus erythematosus and inflammatory bowel disease. From results so far, this approach is able to identify core genes and pathways that are relevant as possible drug targets. An immediate practical application of this approach, where development of a drug has reached clinical stage and the protein targeted by this drug can be identified as a core gene for one or more diseases, is to support decisions on which indications to study in a Phase 2 trial. For instance we have identified the gene that encodes the immune checkpoint receptor PD-1 (programmed death 1) as a core gene for rheumatoid arthritis, systemic lupus erythematosus, and type 1 diabetes. PD-1 agonists that signal through this receptor to down-regulate the immune response have shown effectiveness against rheumatoid arthritis in Phase 2 trials, but have not yet been studied in systemic lupus erythematosus or type 1 diabetes.
Development of likelihood-based methods for inference of causality using pleiotropic genetic instruments.
The relation of C-peptide persistence in Type 1 diabetes to the genetic architecture of Type 1 diabetes and to the risk of short-term and long-term complications of Type 1 diabetes.
Modelling the natural history of diabetic retinopathy, and development of deep learning algorithms for prediction of complications from retinal images
Development of statistical methods for learning to classify with high-dimensional biomarker panels and quantifying the incremental contribution of each biomarker or panel to predictive performance.
Development of statistical and computational methods for pharmacoepidemiologic studies with electronic health records.
From April 2020 to March 2022 much of my time was spent within the Epidemiology Research Cell of Public Health Scotland’s COVID-19 Health Protection Response. In this role I led the design and analysis of REACT-SCOT, a rolling case-control study of all cases of COVID-19 in Scotland since the start of the epidemic with up to ten controls per case, matched for age, sex and general practice and linked to electronic health records. This study was used initially to construct and validate a risk score for severe or fatal COVID-19, and to investigate the relation of severe COVID-19 to drug prescribing. In subsequent work the study was extended to study risk in health care workers and teachers, risk to people designated people as clinically extremely vulnerable and thus eligible for shielding, and nosocomial transmission. Later we studied the efficacy of vaccination in those designated as clinically extremely vulnerable, and the safety of vaccination, focusing on the risk of cerebral venous thrombosis. A list of publications, and links relevant to the “zero COVID” policy that was advocated during 2021-22 is on this page. This paper criticizes the statistical and quantitative modelling methods that were used to make the case for lockdown policies and to quantify the supposed effectiveness of these policies in saving lives: Wood SN, Wit EC, McKeigue PM, Hu D, Flood B, Corcoran L, et al. Some statistical aspects of the Covid-19 response. J R Stat Soc Ser A Stat Soc. 2025;189(1). doi: 10.1093/jrsssa/qnaf092.
Identifying drug targets for type 1 diabetes by genome-wide aggregated trans- effects analysis. McKeigue PM, Colhoun H, Spiliopoulou A. European Foundation for the Study of Diabetes 1/3/26 → 28/2/27.
Real-world pharmacoepidemiology of drugs used in diabetes: harnessing e-health records and artificial intelligence. Colhoun H, McKeigue P. Diabetes UK 2/1/25 → 1/1/2028.
EXCEED - A Pan-European Post-Authorisation Safety Study: Risk of Pancreatic Cancer Among Type 2 Diabetes Patients who Initiated Exenatide as Compared with those who Initiated Other non-Glucagon-Like Peptide 1 Receptor Agonists Colhoun H, McKeigue P. Iqvia Ltd 01/07/21 → 01/05/27.
Association Of C-peptide And Change In C-peptide With Acute And Chronic Complications In Type 1 Diabetes. Colhoun H, McKeigue P. Sanofie Winthrop Industrie 30/04/24 → 30/12/25.
Validation of a new approach for cardiovascular disease risk stratification and therapeutic targeting in T1D. Colhoun H, McKeigue P. Juvenile Diabetes Research Foundation (subcontract from Harvard) 12/02/2024 - 31/10/2025.
Development and Validation of a Transcriptomic-Based Model for Classifying and Predicting Treatment Response in Rheumatoid Arthritis (TRACT-RA) McKeigue P (PI Cos Pitzalis, QMUL). MRC 01/04/21 – 31/03/24.
A UK/Canada collaboration on the genetics of long-term diabetes complications and their risk factors among people with type 1 diabetes. Colhoun H, McKeigue P. MRC 01/04/20 → 31/03/24.
These pages include tutorials, lecture slides and teaching notes prepared when advising colleagues, students and other researchers
Tutorial in the mathematical theory of modelling epidemics while allowing for susceptibility and connectivity to vary between individuals. This theory was first worked out in 2008, but has not been widely disseminated, perhaps because the notation in the original papers is hard to follow. This tutorial presents in an accessible form (level of maths equivalent to a second-year university course) the theory that explains why epidemics in the real world are likely to be smaller than predicted by the classical Kermack-McKendrick model. An article co-written with Simon Wood, discussing the implications of this theory for using models to guide policy in the COVID-19 pandemic has been uploaded as a preprint. Some of this material has been merged into a paper now published in JRSS Series C.
Statistical methods for molecular epidemiology, precision medicine and related areas
Contribution of incorrect statistical methods to the excess of false-positive results in Mendelian randomization analyses. Association for Complex Trait Genetics, March 2025.
Discovery of core genes and drug targets for disease through genome-wide aggregated trans-effects analysis. Novartis Biomedical Research, March 2025.
Most of these publications are available under open access. For those that are not, the final accepted manuscript can usually be found in University of Edinburgh Research Explorer.
McKeigue PM, Iakovliev A, Erabadda B, Colhoun HM, Spiliopoulou A. Genome-Wide Aggregated Trans-Effects Analysis Implicates Deficient Type III Interferon Signaling as a Key Cause of Inflammatory Bowel Disease. Inflamm Bowel Dis. 2025 Nov 1;31(11):3172-3182. doi: 10.1093/ibd/izaf214. PMID: 40996888.
Iakovliev A, Castellini-Pérez O, Erabadda B, Martín J, Barturen G, McKeigue PM, et al. Discovery of core genes for systemic lupus erythematosus via genome-wide aggregated trans-effects analysis. Genes Immun. 2025 Oct;26(5):497-508. doi: 10.1038/s41435-025-00352-4. PMID: 40903491.
Zhou X, Iakovliev A, McGurnaghan S, Erabadda B, Hayward C, McKeigue PM, Spiliopoulou A, Colhoun HM. Genome-Wide Aggregated Trans Effects Analysis for Circulating Proteins Indicates a Key Role of Immune Checkpoints in Type 1 Diabetes. Diabetes. 2025 Oct 1;74(10):1873-1884. doi: 10.2337/db25-0067. PMID: 40788656.
Wood SN, Wit EC, McKeigue PM, Hu D, Flood B, Corcoran L, et al. Some statistical aspects of the Covid-19 response. J R Stat Soc Ser A Stat Soc. 2025;189(1). doi: 10.1093/jrsssa/qnaf092.
Spiliopoulou A, Iakovliev A, Plant D, Sutcliffe M, Sharma S, Cubuk C, et al. Genome-Wide Aggregated Trans Effects Analysis Identifies Genes Encoding Immune Checkpoints as Core Genes for Rheumatoid Arthritis. Arthritis Rheumatol. 2025 Jul;77(7):817-826. doi: 10.1002/art.43125. PMID: 39887658.
Roshandel D, Spiliopoulou A, McGurnaghan SJ, Iakovliev A, Lipschutz D, Hayward C, et al. Genetics of C-Peptide and Age at Diagnosis in Type 1 Diabetes. Diabetes 2025 Feb 1;74(2):223–33.
Mellor J, Kuznetsov D, Heller S, Gall MA, Rosilio M, Amiel SA, et al. Risk factors and prediction of hypoglycaemia using the Hypo-RESOLVE cohort: a secondary analysis of pooled data from insulin clinical trials. Diabetologia. 2024 Aug;67(8):1588–601.
Mellor J, Jiang W, Fleming A, McGurnaghan SJ, Blackbourn LAK, Styles C, et al. Prediction of retinopathy progression using deep learning on retinal images within the Scottish screening programme. Br J Ophthalmol. 2024 May 21;108(6):833–9.
McGurnaghan SJ, McKeigue PM, Blackbourn LAK, Mellor J, Caparrotta TM, Sattar N, et al. Impact of COVID-19 and Non-COVID-19 Hospitalized Pneumonia on Longer-Term Cardiovascular Mortality in People With Type 2 Diabetes: A Nationwide Prospective Cohort Study From Scotland. Diabetes Care. 2024 Aug 1;47(8):1342–9.
Goldmann K, Spiliopoulou A, Iakovliev A, Plant D, Nair N, Cubuk C, et al. Expression quantitative trait loci analysis in rheumatoid arthritis identifies tissue specific variants associated with severity and outcome. Ann Rheum Dis. 2024 Feb 15;83(3):288–99.
Fleming AD, Mellor J, McGurnaghan SJ, Blackbourn LAK, Goatman KA, Styles C, et al. Deep learning detection of diabetic retinopathy in Scotland’s diabetic eye screening programme. Br J Ophthalmol. 2024 Jun 20;108(7):984–8.
Berthon W, McGurnaghan SJ, Blackbourn LAK, Mellor J, Gibb FW, Heller S, et al. Ongoing burden and recent trends in severe hospitalised hypoglycaemia events in people with type 1 and type 2 diabetes in Scotland: A nationwide cohort study 2016-2022. Diabetes Res Clin Pract. 2024 Apr;210:111642.
Berthon W, McGurnaghan SJ, Blackbourn LAK, Bath LE, McAllister DA, Stockton D, et al. Incidence of Type 1 Diabetes in Children Has Fallen to Pre-COVID-19 Pandemic Levels: A Population-Wide Analysis From Scotland. Diabetes Care. 2024 Mar 1;47(3):e26–8.
Fleming AD, Mellor J, McGurnaghan SJ, Blackbourn LAK, Goatman KA, Styles C, et al. Deep learning detection of diabetic retinopathy in Scotland’s diabetic eye screening programme. Br J Ophthalmol. 2023 Sep 13;bjo-2023-323395.
Mellor J, Jiang W, Fleming A, McGurnaghan SJ, Blackbourn L, Styles C, Storkey AJ, McKeigue PM, Colhoun HM; Scottish Diabetes Research Network Epidemiology Group. Can deep learning on retinal images augment known risk factors for cardiovascular disease prediction in diabetes? A prospective cohort study from the national screening programme in Scotland. Int J Med Inform. 2023 Jul;175:105072. doi: 10.1016/j.ijmedinf.2023.105072. Epub 2023 Apr 18. PMID: 37167840.
Iakovliev A, McGurnaghan SJ, Hayward C, Colombo M, Lipschutz D, Spiliopoulou A, Colhoun HM, McKeigue PM. Genome-wide aggregated trans-effects on risk of type 1 diabetes: A test of the “omnigenic” sparse effector hypothesis of complex trait genetics. Am J Hum Genet. 2023 Jun 1;110(6):913-926. doi: 10.1016/j.ajhg.2023.04.003. Epub 2023 May 9. PMID: 37164005; PMCID: PMC10257008.
Wang H, Cordiner RLM, Huang Y, Donnelly L, Hapca S, Collier A, McKnight J, Kennon B, Gibb F, McKeigue P, Wild SH, Colhoun H, Chalmers J, Petrie J, Sattar N, MacDonald T, McCrimmon RJ, Morales DR, Pearson ER; Scottish Diabetes Research Network Epidemiology Group. Cardiovascular Safety in Type 2 Diabetes With Sulfonylureas as Second-line Drugs: A Nationwide Population-Based Comparative Safety Study. Diabetes Care. 2023 May 1;46(5):967-977. doi: 10.2337/dc22-1238. PMID: 36944118; PMCID: PMC10154665.
McGurnaghan SJ, Blackbourn LAK, Caparrotta TM, Mellor J, Barnett A, Collier A, Sattar N, McKnight J, Petrie J, Philip S, Lindsay R, Hughes K, McAllister D, Leese GP, Pearson ER, Wild S, McKeigue PM, Colhoun HM. Cohort profile: the Scottish Diabetes Research Network national diabetes cohort - a population-based cohort of people with diabetes in Scotland. BMJ Open. 2022 Oct 12;12(10):e063046. doi: 10.1136/bmjopen-2022-063046. PMID: 36223968; PMCID: PMC9562713.
Höhn A, McGurnaghan SJ, Caparrotta TM, Jeyam A, O’Reilly JE, Blackbourn LAK, Hatam S, Dudel C, Seaman RJ, Mellor J, Sattar N, McCrimmon RJ, Kennon B, Petrie JR, Wild S, McKeigue PM, Colhoun HM; SDRN-Epi Group. Large socioeconomic gap in period life expectancy and life years spent with complications of diabetes in the Scottish population with type 1 diabetes, 2013-2018. PLoS One. 2022 Aug 11;17(8):e0271110. doi: 10.1371/journal.pone.0271110. PMID: 35951518; PMCID: PMC9371295.
Paul M McKeigue, Stuart McGurnaghan, Luke Blackbourn, Louise E Bath, David A McAllister, Thomas M Caparrotta, Sarah H Wild, Simon N Wood, Diane Stockton, Helen M Colhoun. Relation of incident Type 1 diabetes to recent COVID-19 infection: cohort study using e-health record linkage in Scotland. Diabetes Care 2022 doi: 10.2337/dc22-0385
McKeigue P. Fitting joint models of longitudinal observations and time to event by sequential Bayesian updating. Stat Methods Med Res. 2022 May 31:9622802221104241. doi: 10.1177/09622802221104241. Epub ahead of print. PMID: 35642267.
Rivellese F, Surace AEA, Goldmann K, Sciacca E, Çubuk C, Giorli G, John CR, Nerviani A, Fossati-Jimack L, Thorborn G, Ahmed M, Prediletto E, Church SE, Hudson BM, Warren SE, McKeigue PM, Humby F, Bombardieri M, Barnes MR, Lewis MJ, Pitzalis C; R4RA collaborative group. Rituximab versus tocilizumab in rheumatoid arthritis: synovial biopsy-based biomarker analysis of the phase 4 R4RA randomized trial. Nat Med. 2022 Jun;28(6):1256-1268. doi: 10.1038/s41591-022-01789-0. Epub 2022 May 19. PMID: 35589854; PMCID: PMC9205785.
Paul M McKeigue, Duncan Porter, Rosemary J Hollick, Stuart H Ralston, David A McAllister, Helen M Colhoun. Risk of severe COVID-19 in patients with inflammatory rheumatic diseases treated with immunosuppressive therapy in Scotland. Scand J Rheumatol. 2022 May 12:1-6. doi: 10.1080/03009742.2022.2063376
McKeigue PM, McAllister DA, Hutchinson SJ, Robertson C, Stockton D, Colhoun HM, et al. Efficacy of vaccination against severe COVID-19 in relation to Delta variant and time since second dose: the REACT-SCOT case-control study. Lancet Respir Med. 2022 Jun;10(6):566-572. doi: 10.1016/S2213-2600(22)00045-5. Epub 2022 Feb 25.
McKeigue PM, Burgul R, Bishop J, Robertson C, McMenamin J, O’Leary M, McAllister DA, Colhoun HM. Association of cerebral venous thrombosis with recent COVID-19 vaccination: case-crossover study using ascertainment through neuroimaging in Scotland. BMC Infect Dis. 2021 Dec 20;21(1):1275. doi: 10.1186/s12879-021-06960-5. PMID: 34930153; PMCID: PMC8686098.
Jeyam A, Gibb FW, McKnight JA, O’Reilly JE, Caparrotta TM, Höhn A, McGurnaghan SJ, Blackbourn LAK, Hatam S, Kennon B, McCrimmon RJ, Leese G, Philip S, Sattar N, McKeigue PM, Colhoun HM; Scottish Diabetes Research Network Epidemiology Group. Flash monitor initiation is associated with improvements in HbA1c levels and DKA rates among people with type 1 diabetes in Scotland: a retrospective nationwide observational study. Diabetologia. 2021 Oct 7. doi: 10.1007/s00125-021-05578-1. Epub ahead of print. PMID: 34618177.
Shah ASV, Gribben C, Bishop J, Hanlon P, Caldwell D, Wood R, Reid M, McMenamin J, Goldberg D, Stockton D, Hutchinson S, Robertson C, McKeigue PM, Colhoun HM, McAllister DA. Effect of Vaccination on Transmission of SARS-CoV-2. N Engl J Med. 2021 Sep 8. doi: 10.1056/NEJMc2106757. Epub ahead of print. PMID: 34496200.
Fenton L, Gribben C, Caldwell D, Colville S, Bishop J, Reid M, White J, Campbell M, Hutchinson S, Robertson C, Colhoun HM, Wood R, McKeigue PM, McAllister DA. Risk of hospital admission with covid-19 among teachers compared with healthcare workers and other adults of working age in Scotland, March 2020 to July 2021: population based case-control study. BMJ. 2021 Sep 1;374:n2060. doi: 10.1136/bmj.n2060. PMID: 34470747; PMCID: PMC8408959.
O’Reilly JE, Jeyam A, Caparrotta TM, Mellor J, Hohn A, McKeigue PM, McGurnaghan SJ, Blackbourn LAK, McCrimmon R, Wild SH, Petrie JR, McKnight JA, Kennon B, Chalmers J, Phillip S, Leese G, Lindsay RS, Sattar N, Gibb FW, Colhoun HM; Scottish Diabetes Research Network Epidemiology Group. Rising Rates and Widening Socioeconomic Disparities in Diabetic Ketoacidosis in Type 1 Diabetes in Scotland: A Nationwide Retrospective Cohort Observational Study. Diabetes Care. 2021 Jul 8:dc210689. doi: 10.2337/dc21-0689. Epub ahead of print. PMID: 34244330.
McGurnaghan SJ, McKeigue PM, Read SH, Franzen S, Svensson AM, Colombo M, Livingstone S, Farran B, Caparrotta TM, Blackbourn LAK, Mellor J, Thoma I, Sattar N, Wild SH, Gudbjörnsdottir S, Colhoun HM; Swedish National Diabetes Register and the Scottish Diabetes Research Network Epidemiology Group. Development and validation of a cardiovascular risk prediction model in type 1 diabetes. Diabetologia. 2021 Jun 9. doi: 10.1007/s00125-021-05478-4. Epub ahead of print. PMID: 34106282.
Jeyam A, Gibb FW, McKnight JA, Kennon B, O’Reilly JE, Caparrotta TM, Höhn A, McGurnaghan SJ, Blackbourn LAK, Hatam S, McCrimmon RJ, Leese G, Lindsay RS, Petrie J, Chalmers J, Philip S, Wild SH, Sattar N, McKeigue PM, Colhoun HM; Scottish Diabetes Research Network (SDRN) Epidemiology Group. Marked improvements in glycaemic outcomes following insulin pump therapy initiation in people with type 1 diabetes: a nationwide observational study in Scotland. Diabetologia. 2021 Mar 8. doi: 10.1007/s00125-021-05413-7. Epub ahead of print. PMID: 33686483.
Shah ASV, Gribben C, Bishop J, Hanlon P, Caldwell D, Wood R, Reid M, McMenamin J, Goldberg D, Stockton D, Hutchinson S, Robertson C, McKeigue PM, Colhoun HM, McAllister DA. Effect of Vaccination on Transmission of SARS-CoV-2. N Engl J Med. 2021 Sep 8:NEJMc2106757. doi: 10.1056/NEJMc2106757. Epub ahead of print. PMID: 34496200; PMCID: PMC8451182.
McKeigue, P.M., McAllister, D.A., Caldwell, D. et al. Relation of severe COVID-19 in Scotland to transmission-related factors and risk conditions eligible for shielding support: REACT-SCOT case-control study. BMC Med 19, 149 (2021). https://doi.org/10.1186/s12916-021-02021-5
Wood R, Thomson E, Galbraith R, Gribben C, Caldwell D, Bishop J, Reid M, Shah ASV, Templeton K, Goldberg D, Robertson C, Hutchinson SJ, Colhoun HM, McKeigue PM, McAllister DA. Sharing a household with children and risk of COVID-19: a study of over 300 000 adults living in healthcare worker households in Scotland. Arch Dis Child. 2021 Mar 18:archdischild-2021-321604. doi: 10.1136/archdischild-2021-321604. Epub ahead of print. PMID: 33737319.
McKeigue PM, Kennedy S, Weir A, Bishop J, McGurnaghan SJ, McAllister D, Robertson C, Wood R, Lone N, Murray J, Caparrotta TM, Smith-Palmer A, Goldberg D, McMenamin J, Guthrie B, Hutchinson S, Colhoun HM; Public Health Scotland COVID-19 Health Protection Study Group. Relation of severe COVID-19 to polypharmacy and prescribing of psychotropic drugs: the REACT-SCOT case-control study. BMC Med. 2021 Feb 22;19(1):51. doi: 10.1186/s12916-021-01907-8. PMID: 33612113.
Höhn A, Jeyam A, Caparrotta TM, McGurnaghan SJ, O’Reilly JE, Blackbourn LAK, McCrimmon RJ, Leese GP, McKnight JA, Kennon B, Lindsay RS, Sattar N, Wild SH, McKeigue PM, Colhoun HM; Scottish Diabetes Research Network (SDRN) Epidemiology Group. The association of polypharmacy and high-risk drug classes with adverse health outcomes in the Scottish population with type 1 diabetes. Diabetologia. 2021 Feb 19. doi: 10.1007/s00125-021-05394-7. Epub ahead of print. PMID: 33608768.
McGurnaghan SJ, Weir A, Bishop J, Kennedy S, Blackbourn LAK, McAllister DA, Hutchinson S, Caparrotta TM, Mellor J, Jeyam A, O’Reilly JE, Wild SH, Hatam S, Höhn A, Colombo M, Robertson C, Lone N, Murray J, Butterly E, Petrie J, Kennon B, McCrimmon R, Lindsay R, Pearson E, Sattar N, McKnight J, Philip S, Collier A, McMenamin J, Smith-Palmer A, Goldberg D, McKeigue PM, Colhoun HM; Public Health Scotland COVID-19 Health Protection Study Group; Scottish Diabetes Research Network Epidemiology Group. Risks of and risk factors for COVID-19 disease in people with diabetes: a cohort study of the total population of Scotland. Lancet Diabetes Endocrinol. 2021 Feb;9(2):82-93. doi: 10.1016/S2213-8587(20)30405-8. Epub 2020 Dec 23. PMID: 33357491; PMCID: PMC7832778.
Jeyam A, Colhoun H, McGurnaghan S, Blackbourn L, McDonald TJ, Palmer CNA, McKnight JA, Strachan MWJ, Patrick AW, Chalmers J, Lindsay RS, Petrie JR, Thekkepat S, Collier A, MacRury S, McKeigue PM; SDRNT1BIO Investigators. Clinical Impact of Residual C-Peptide Secretion in Type 1 Diabetes on Glycemia and Microvascular Complications. Diabetes Care. 2020 Dec 10:dc200567. doi: 10.2337/dc20-0567. Epub ahead of print. PMID: 33303639.
Colombo M, Shehni AA, Thoma I, McGurnaghan SJ, Blackbourn LAK, Wilkinson H, Collier A, Patrick AW, Petrie JR, McKeigue PM, Saldova R, Colhoun HM. Quantitative levels of serum N-glycans in type 1 diabetes and their association with kidney disease. Glycobiology. 2020 Nov 27:cwaa106. doi: 10.1093/glycob/cwaa106. Epub ahead of print. PMID: 33245334.
Shah ASV, Wood R, Gribben C, Caldwell D, Bishop J, Weir A, Kennedy S, Reid M, Smith-Palmer A, Goldberg D, McMenamin J, Fischbacher C, Robertson C, Hutchinson S, McKeigue P, Colhoun H, McAllister DA. Risk of hospital admission with coronavirus disease 2019 in healthcare workers and their households: nationwide linkage cohort study. BMJ. 2020 Oct 28;371:m3582. doi: 10.1136/bmj.m3582. PMID: 33115726; PMCID: PMC7591828.
McKeigue PM, Weir A, Bishop J, McGurnaghan SJ, Kennedy S, McAllister D, Robertson C, Wood R, Lone N, Murray J, Caparrotta TM, Smith-Palmer A, Goldberg D, McMenamin J, Ramsay C, Hutchinson S, Colhoun HM; Public Health Scotland COVID-19 Health Protection Study Group. Rapid Epidemiological Analysis of Comorbidities and Treatments as risk factors for COVID-19 in Scotland (REACT- SCOT): A population-based case-control study. PLoS Med. 2020 Oct 20;17(10):e1003374. doi: 10.1371/journal.pmed.1003374. PMID: 33079969; PMCID: PMC7575101.
Štambuk J, Nakić N, Vučković F, Pučić-Baković M, Razdorov G, Trbojević- Akmačić I, Novokmet M, Keser T, Vilaj M, Štambuk T, Gudelj I, Šimurina M, Song M, Wang H, Salihović MP, Campbell H, Rudan I, Kolčić I, Eller LA, McKeigue P, Robb ML, Halfvarson J, Kurtoglu M, Annese V, Škarić-Jurić T, Molokhia M, Polašek O, Hayward C, Kibuuka H, Thaqi K, Primorac D, Gieger C, Nitayaphan S, Spector T, Wang Y, Tillin T, Chaturvedi N, Wilson JF, Schanfield M, Filipenko M, Wang W, Lauc G. Global variability of the human IgG glycome. Aging (Albany NY). 2020 Aug 12;12(15):15222-15259. doi: 10.18632/aging.103884. Epub 2020 Aug 12. PMID: 32788422; PMCID: PMC7467356.
Marcovecchio ML, Colombo M, Dalton RN, McKeigue PM, Benitez-Aguirre P, Cameron FJ, Chiesa ST, Couper JJ, Craig ME, Daneman D, Davis EA, Deanfield JE, Donaghue KC, Jones TW, Mahmud FH, Marshall SM, Neil A, Colhoun HM, Dunger DB; AdDIT and the SDRNT1BIO Investigators. Biomarkers associated with early stages of kidney disease in adolescents with type 1 diabetes. Pediatr Diabetes. 2020 Aug 11. doi: 10.1111/pedi.13095. Epub ahead of print. PMID: 32783254.
Cherlin S, Lewis MJ, Plant D, Nair N, Goldmann K, Tzanis E, Barnes MR, McKeigue P, Barrett JH, Pitzalis C, Barton A; MATURA Consortium, Cordell HJ. Investigation of genetically regulated gene expression and response to treatment in rheumatoid arthritis highlights an association between IL18RAP expression and treatment response. Ann Rheum Dis. 2020 Nov;79(11):1446-1452. doi: 10.1136/annrheumdis-2020-217204. Epub 2020 Jul 30. PMID: 32732242; PMCID: PMC7569378.
Barbu MC, Spiliopoulou A, Colombo M, McKeigue P, Clarke TK, Howard DM, Adams MJ, Shen X, Lawrie SM, McIntosh AM, Whalley HC. Expression quantitative trait loci-derived scores and white matter microstructure in UK Biobank: a novel approach to integrating genetics and neuroimaging. Transl Psychiatry. 2020 Feb 4;10(1):55. doi: 10.1038/s41398-020-0724-y. PMID: 32066731; PMCID: PMC7026054.
O'Reilly JE, Blackbourn LAK, Caparrotta TM, Jeyam A, Kennon B, Leese GP, Lindsay RS, McCrimmon RJ, McGurnaghan SJ, McKeigue PM, McKnight JA, Petrie JR, Philip S, Sattar N, Wild SH, Colhoun HM; Scottish Diabetes Research Network Epidemiology Group. Time trends in deaths before age 50 years in people with type 1 diabetes: a nationwide analysis from Scotland 2004-2017. Diabetologia. 2020 Aug;63(8):1626-1636. doi: 10.1007/s00125-020-05173-w. Epub 2020 May 26. PMID: 32451572; PMCID: PMC7351819.
Caparrotta TM, Blackbourn LAK, McGurnaghan SJ, Chalmers J, Lindsay R, McCrimmon R, McKnight J, Wild S, Petrie JR, Philip S, McKeigue PM, Webb DJ, Sattar N, Colhoun HM; Scottish Diabetes Research Network–Epidemiology Group. Prescribing Paradigm Shift? Applying the 2019 European Society of Cardiology-Led Guidelines on Diabetes, Prediabetes, and Cardiovascular Disease to Assess Eligibility for Sodium-Glucose Cotransporter 2 Inhibitors or Glucagon-Like Peptide 1 Receptor Agonists as First-Line Monotherapy (or Add-on to Metformin Monotherapy) in Type 2 Diabetes in Scotland. Diabetes Care. 2020 Jun 24:dc200120. doi: 10.2337/dc20-0120. Epub ahead of print. PMID: 32581068.
Li X, Timofeeva M, Spiliopoulou A, McKeigue P, He Y, Zhang X, Svinti V, Campbell H, Houlston RS, Tomlinson IPM, Farrington SM, Dunlop MG, Theodoratou E. Prediction of colorectal cancer risk based on profiling with common genetic variants. Int J Cancer. 2020 Jul 7. doi: 10.1002/ijc.33191. Epub ahead of print. PMID: 32638365.
Jeyam A, McGurnaghan SJ, Blackbourn LAK, McKnight JM, Green F, Collier A, McKeigue PM, Colhoun HM; SDRNT1BIO Investigators. Diabetic Neuropathy Is a Substantial Burden in People With Type 1 Diabetes and Is Strongly Associated With Socioeconomic Disadvantage: A Population-Representative Study From Scotland. Diabetes Care. 2020 Jan 23:dc191582. doi: 10.2337/dc19-1582. Epub ahead of print. PMID: 31974100.
Colombo M, McGurnaghan SJ, Blackbourn LAK, Dalton RN, Dunger D, Bell S, Petrie JR, Green F, MacRury S, McKnight JA, Chalmers J, Collier A, McKeigue PM, Colhoun HM; Scottish Diabetes Research Network (SDRN) Type 1 Bioresource Investigators. Comparison of serum and urinary biomarker panels with albumin/creatinine ratio in the prediction of renal function decline in type 1 diabetes. Diabetologia. 2020 Jan 8:10.1007/s00125-019-05081-8. doi: 10.1007/s00125-019-05081-8. Epub ahead of print. PMID: 31915892.
Colombo M, McGurnaghan SJ, Bell S, MacKenzie F, Patrick AW, Petrie JR, McKnight JA, MacRury S, Traynor J, Metcalfe W, McKeigue PM, Colhoun HM; Scottish Diabetes Research Network (SDRN) Type 1 Bioresource Investigators and the Scottish Renal Registry. Predicting renal disease progression in a large contemporary cohort with type 1 diabetes mellitus. Diabetologia. 2020 Mar;63(3):636-647. doi: 10.1007/s00125-019-05052-z. Epub 2019 Dec 5. PMID: 31807796.
Li X, Meng X, He Y, Spiliopoulou A, Timofeeva M, Wei WQ, Gifford A, Yang T, Varley T, Tzoulaki I, Joshi P, Denny JC, Mckeigue P, Campbell H, Theodoratou E. Genetically determined serum urate levels and cardiovascular and other diseases in UK Biobank cohort: A phenome-wide mendelian randomization study. PLoS Med. 2019 Oct 18;16(10):e1002937. doi: 10.1371/journal.pmed.1002937. PMID: 31626644; PMCID: PMC6799886.
Ochs A, McGurnaghan S, Black MW, Leese GP, Philip S, Sattar N, Styles C, Wild SH, McKeigue PM, Colhoun HM; Scottish Diabetes Research Network Epidemiology Group and the Diabetic Retinopathy Screening Collaborative. Use of personalised risk-based screening schedules to optimise workload and sojourn time in screening programmes for diabetic retinopathy: A retrospective cohort study. PLoS Med. 2019 Oct 17;16(10):e1002945. doi: 10.1371/journal.pmed.1002945. PMID: 31622334; PMCID: PMC6797087.
Meng X, Li X, Timofeeva MN, He Y, Spiliopoulou A, Wei WQ, Gifford A, Wu H, Varley T, Joshi P, Denny JC, Farrington SM, Zgaga L, Dunlop MG, McKeigue P, Campbell H, Theodoratou E. Phenome-wide Mendelian-randomization study of genetically determined vitamin D on multiple health outcomes using the UK Biobank study. Int J Epidemiol. 2019 Oct 1;48(5):1425-1434. doi: 10.1093/ije/dyz182. PMID: 31518429; PMCID: PMC6857754.
Salem RM, Todd JN, Sandholm N, Cole JB, Chen WM, Andrews D, Pezzolesi MG, McKeigue PM, Hiraki LT, Qiu C, Nair V, Di Liao C, Cao JJ, Valo E, Onengut- Gumuscu S, Smiles AM, McGurnaghan SJ, Haukka JK, Harjutsalo V, Brennan EP, van Zuydam N, Ahlqvist E, Doyle R, Ahluwalia TS, Lajer M, Hughes MF, Park J, Skupien J, Spiliopoulou A, Liu A, Menon R, Boustany-Kari CM, Kang HM, Nelson RG, Klein R, Klein BE, Lee KE, Gao X, Mauer M, Maestroni S, Caramori ML, de Boer IH, Miller RG, Guo J, Boright AP, Tregouet D, Gyorgy B, Snell-Bergeon JK, Maahs DM, Bull SB, Canty AJ, Palmer CNA, Stechemesser L, Paulweber B, Weitgasser R, Sokolovska J, Rovīte V, Pīrāgs V, Prakapiene E, Radzeviciene L, Verkauskiene R, Panduru NM, Groop LC, McCarthy MI, Gu HF, Möllsten A, Falhammar H, Brismar K, Martin F, Rossing P, Costacou T, Zerbini G, Marre M, Hadjadj S, McKnight AJ, Forsblom C, McKay G, Godson C, Maxwell AP, Kretzler M, Susztak K, Colhoun HM, Krolewski A, Paterson AD, Groop PH, Rich SS, Hirschhorn JN, Florez JC; SUMMIT Consortium, DCCT/EDIC Research Group, GENIE Consortium. Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Glomerular Basement Membrane Collagen. J Am Soc Nephrol. 2019 Oct;30(10):2000-2016. doi: 10.1681/ASN.2019030218. Epub 2019 Sep 19. PMID: 31537649; PMCID: PMC6779358.
McKeigue PM, Spiliopoulou A, McGurnaghan S, Colombo M, Blackbourn L, McDonald TJ, Onengut-Gomuscu S, Rich SS, A Palmer CN, McKnight JA, J Strachan MW, Patrick AW, Chalmers J, Lindsay RS, Petrie JR, Thekkepat S, Collier A, MacRury S, Colhoun HM. Persistent C-peptide secretion in Type 1 diabetes and its relationship to the genetic architecture of diabetes. BMC Med. 2019 Aug 23;17(1):165. doi: 10.1186/s12916-019-1392-8. PubMed PMID: 31438962.
Floyd JS, Bloch KM, Brody JA, Maroteau C, Siddiqui MK, Gregory R, Carr DF, Molokhia M, Liu X, Bis JC, Ahmed A, Liu X, Hallberg P, Yue QY, Magnusson PKE, Brisson D, Wiggins KL, Morrison AC, Khoury E, McKeigue P, Stricker BH, Lapeyre-Mestre M, Heckbert SR, Gallagher AM, Chinoy H, Gibbs RA, Bondon-Guitton E, Tracy R, Boerwinkle E, Gaudet D, Conforti A, van Staa T, Sitlani CM, Rice KM, aitland-van der Zee AH, Wadelius M, Morris AP, Pirmohamed M, Palmer CAN, Psaty BM, Alfirevic A; PREDICTION-ADR Consortium and EUDRAGENE. Pharmacogenomics of statin-related myopathy: Meta-analysis of rare variants from whole-exome sequencing. PLoS One. 2019 Jun 26;14(6):e0218115. doi: 10.1371/journal.pone.0218115. eCollection 2019. PubMed PMID: 31242253; PubMed Central PMCID: PMC6594672.
Colombo M, Valo E, McGurnaghan SJ, Sandholm N, Blackbourn LAK, Dalton RN, Dunger D, Groop PH, McKeigue PM, Forsblom C, Colhoun HM; FinnDiane Study Group and the Scottish Diabetes Research Network (SDRN) Type 1 Bioresource Collaboration. Biomarker panels associated with progression of renal disease in type 1 diabetes. Diabetologia. 2019 Jun 20. doi: 10.1007/s00125-019-4915-0. [Epub]
Mair C, Wulaningsih W, Jeyam A, McGurnaghan S, Blackbourn L, Kennon B, Leese G, Lindsay R, McCrimmon RJ, McKnight J, Petrie JR, Sattar N, Wild SH, Conway N, Craigie I, Robertson K, Bath L, McKeigue PM, Colhoun HM; Scottish Diabetes Research Network (SDRN) Epidemiology Group. Glycaemic control trends in people with type 1 diabetes in Scotland 2004-2016. Diabetologia. 2019 Aug;62(8):1375-1384. doi: 10.1007/s00125-019-4900-7. Epub 2019 May 18. PubMed PMID: 31104095.
Spiliopoulou A, Colombo M, Plant D, Nair N, Cui J, Coenen MJ, Ikari K, Yamanaka H, Saevarsdottir S, Padyukov L, Bridges SL Jr, Kimberly RP, Okada Y, van Riel PLC, Wolbink G, van der Horst-Bruinsma IE, de Vries N, Tak PP, Ohmura K, Canhão H, Guchelaar HJ, Huizinga TW, Criswell LA, Raychaudhuri S, Weinblatt ME, Wilson AG, Mariette X, Isaacs JD, Morgan AW, Pitzalis C, Barton A, McKeigue P. Association of response to TNF inhibitors in rheumatoid arthritis with quantitative trait loci for CD40 and CD39. Ann Rheum Dis. 2019 Apr 29. doi: 10.1136/annrheumdis-2018-214877. [Epub ahead of print] PubMed PMID: 31036624.
Hensor EMA, McKeigue P, Ling SF, Colombo M, Barrett JH, Nam JL, Freeston J, Buch MH, Spiliopoulou A, Agakov F, Kelly S, Lewis MJ, Verstappen SMM, MacGregor AJ, Viatte S, Barton A, Pitzalis C, Emery P, Conaghan PG, Morgan AW. Validity of a two-component imaging-derived disease activity score for improved assessment of synovitis in early rheumatoid arthritis. Rheumatology (Oxford). 2019 Mar 1. doi: 10.1093/rheumatology/kez049. [Epub ahead of print] PubMed PMID: 30824919.
McGurnaghan SJ, Brierley L, Caparrotta TM, McKeigue PM, Blackbourn LAK, Wild SH, Leese GP, McCrimmon RJ, McKnight JA, Pearson ER, Petrie JR, Sattar N, Colhoun HM; Scottish Diabetes Research Network Epidemiology Group. The effect of dapagliflozin on glycaemic control and other cardiovascular disease risk factors in type 2 diabetes mellitus: a real-world observational study. Diabetologia. 2019 Apr;62(4):621-632. doi: 10.1007/s00125-018-4806-9. Epub 2019 Jan 10. PubMed PMID: 30631892.
Colombo M, Looker HC, Farran B, Hess S, Groop L, Palmer CNA, Brosnan MJ,Dalton RN, Wong M, Turner C, Ahlqvist E, Dunger D, Agakov F, Durrington P,ivingstone S, Betteridge J, McKeigue PM, Colhoun HM; SUMMIT Investigators. Serum kidney injury molecule 1 and β(2)-microglobulin perform as well as larger biomarker panels for prediction of rapid decline in renal function in type 2 diabetes. Diabetologia. 2018 Oct 5. doi: 10.1007/s00125-018-4741-9. [Epub ah.pead of print] PubMed PMID: 30288572.
Cherlin S, Plant D , Taylor JC, Colombo M, Spiliopoulou A , Tzanis E , Morgan AW, Barnes MR , McKeigue P, Barrett JH, Pitzalis C, Barton A, MATURA Consortium, Cordell HJ. Prediction of treatment response in rheumatoid arthritis patients using genome-wide SNP data. Genetic Epidemiology 2018: Dec;42(8):754-771. doi: 10.1002/gepi.22159
Massey J, Plant D, Hyrich K, Morgan AW, Wilson AG, Spiliopoulou A, Colombo M, McKeigue P, Isaacs J, Cordell H, Pitzalis C, Barton A; BRAGGSS, MATURA Consortium. Genome-wide association study of response to tumour necrosis factor inhibitor therapy in rheumatoid arthritis. Pharmacogenomics J. 2018 Aug 31. doi: 10.1038/s41397-018-0040-6. [Epub ahead of print] PubMed PMID: 30166627.
McKeigue P. Quantifying performance of a diagnostic test as the expected information for discrimination: Relation to the C-statistic. Stat Methods Med Res. 2018 Jan 1:962280218776989. doi: 10.1177/0962280218776989. [Epub ahead of print] PubMed PMID: 29978758.
Colombo M,, Looker HC, Farran B, Agakov F, Brosnan MJ, Welsh P, Sattar N, Livingston SJ, Durrington PN, Betteridge DJ, McKeigue PM, Colhoun HM. Apolipoprotein CIII and N-Terminal Prohormone B-type Natriuretic Peptide as Independent Predictors for Cardiovascular Disease in Type 2 Diabetes. Atherosclerosis 2018, May 21;274:182-190
McKeigue P. Sample size requirements for learning to classify with high-dimensional biomarker panels. Stat Methods Med Res. 2018 Jan 1:962280217738807. doi: 10.1177/0962280217738807. [Epub ahead of print] PubMed PMID: 29179643
jmseq: Joint
modelling of longitudinal data and time to eventFitting joint models of longitudinal data and time to event by sequential Bayesian updating (work in progress).
hsnpyr:
Regularized horseshoe regression in PythonA Python package for fitting regularized horseshoe logistic
regression models using NumPyro, designed for predicting outcomes from
high-dimensional biomarker panels. It provides K-fold cross-validation,
projection predictive variable selection, learning curves, and
evaluation metrics including the C-statistic and expected information
for discrimination. This is a reimplementation with computational
advantages over the R package hsstan described below.
hsstan:
Hierarchical Shrinkage Stan Models for Biomarker SelectionDevelopers: Marco Colombo, Paul McKeigue, Athina Spiliopoulou. Linear and logistic regression models penalized with hierarchical shrinkage priors for selection of biomarkers (or more general variable selection), which can be fitted using Stan. It implements the horseshoe and regularized horseshoe priors Piironen and Vehtari 2017, as well as the projection predictive selection approach to recover a sparse set of predictive biomarkers (Piironen, Paasiniemi and Vehtari (2020) doi:10.1214/20-EJS1711).
wevid: Weight
of evidence for evaluating a classifierAn R package for computing distributions of the weight of evidence (log Bayes factor) favouring case over noncase status, to quantify the performance of a binary classifier. Given prior probabilities, posterior probabilities, and observed case-control status, it computes the expected information for discrimination and plots the distributions of weight of evidence in cases and controls. In comparison with the C-statistic (area under ROC curve), the expected weight of evidence has several advantages as a summary measure of predictive performance.
To calculate the required sample size for learning to predict from high-dimensional biomarker panels, classical statistical power calculations are not very useful. What researchers really need is a learning curve, showing how the expected predictive performance of the trained model depends on the size of the training sample from which this model is learned.
I have described a simple method for calculating the sample size required to learn to classify with a high-dimensional biomarker panel, based on the asymptotic distribution of the weight of evidence (log Bayes factor) in a recent paper: Sample size requirements for learning to classify with high-dimensional biomarker panels (Statistical Methods for Medical Research 2017, final version now on PubMed). The assumptions underlying this method are:-
There are no redundant biomarkers in the sense that none of them can be calculated as weighted sums of the others (covariance matrix is of full rank)
The effects of the biomarkers can be approximated by a linear discriminant model in which the class-conditional distributions of the biomarkers are gaussian with the same covariance in cases and noncases.
The class-conditional correlations between the biomarkers are the same as the correlations between their prior effect sizes.
The method is implemented in an online sample size calculator, written with the R shiny package and deployed at https://pmckeigue.shinyapps.io/sampsizeapp/.
To use it, move the sliders to specify:-
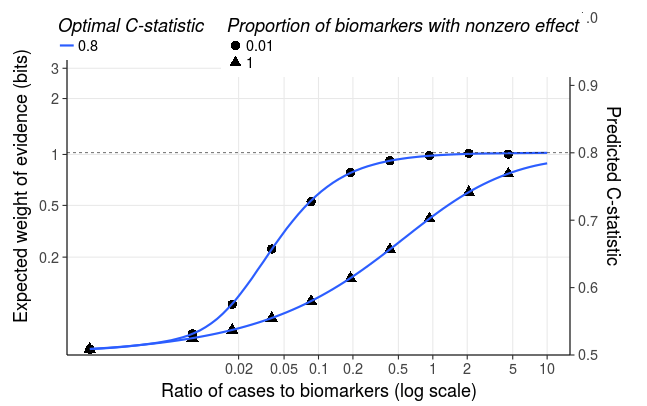
the performance of the optimal classifier that could be learned from a training sample of infinite size. This is specified as the C-statistic (area under the ROC curve). C-statistics of 0.80, 0.88 and 0.925 are equivalent to expected information for discrimination of 1, 2 and 3 bits respectively.
The proportion of biomarkers that have nonzero effect sizes, based on a spike-and-slab mixture model for the distribution of effect sizes. For instance, specifying 0.1 is equivalent to specifying that 90% of biomarkers have zero effect size (the spike component), and the remaining 10% have a gaussian distribution of effect sizes with mean zero (the slab component). The app will plot the learning curve based on the proportion that you specify, and also based on a model in which the biomarkers have a gaussian distribution of effect sizes. If the biomarkers in your panel are unselected (for instance a genome-wide profile of gene transcription levels) you may expect that only a small proportion of these biomarkers contain predictive information. If your biomarker panel has been preselected for relevance (for instance markers previously reported to be associated with the outcome under study, you may expect the proportion with nonzero effects to be relatively high.
Then click the “Submit” button
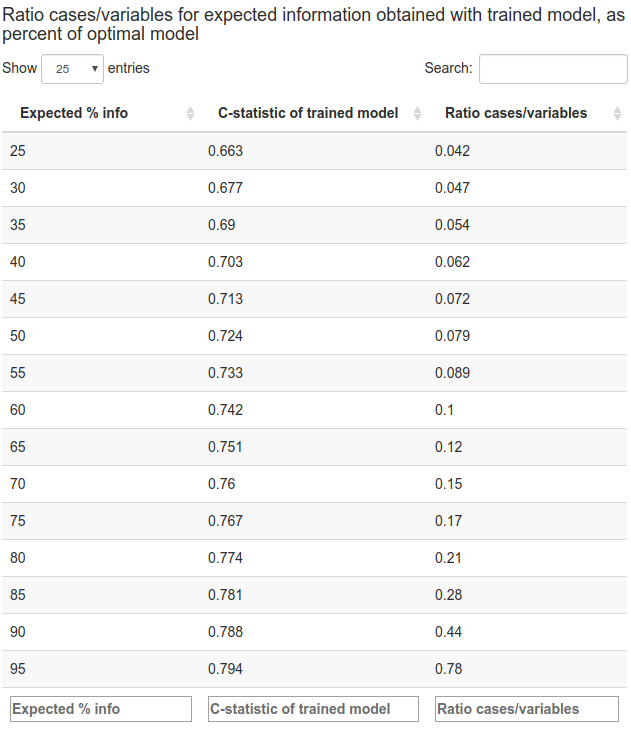
The table generated by the app has three columns:
the expectation of the information for discrimination extracted by the trained model, as a percentage of the information for discrimination extracted by the optimal model that would be learned from a sample of infinite size. This is scaled from 25% to 90%.
the C-statistic corresponding to these values of expected information for discrimination.
the ratio of cases to variables, assuming a balanced study design with equal numbers of cases and controls.
The app also plots a learning curve, showing how the expected information for discrimination and C-statistic obtained with the trained model depends on the ratio of cases to variables. In this example, where the optimal model has an expected discrimination of 1 bit equivalent to a C-statistic of 0.80, and 1% of biomarkers have nonzero effects, a sample size of at least 0.1 cases per variable is required to learn a model that has expected information for discrimination 60% of that obtained with the optimal model.